Code & Dataset
Brain Transcriptome Single-cell (BTS) Atlas
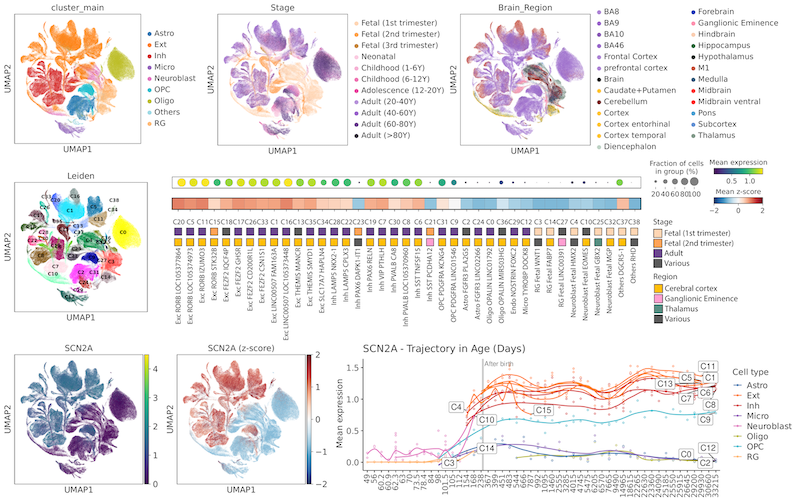
Brain Transcriptome Single-cell Atlas (BTS) Anndata, Seurat object, and Celltypist model for further use of the atlas. The Celltypist model can be utilized to accurately annotate cell types in new datasets based on the atlas. Plots illustrating the expression profile for 3,380 neurological disorder risk genes across the atlas are also uploaded. Further availability for the data can be requested by the corresponding author.
This dataset is published in Kim, S., Lee, J., Koh, I.G. et al. An integrative single-cell atlas for exploring the cellular and temporal specificity of genes related to neurological disorders during human brain development. Exp Mol Med 56, 2271–2282 (2024). https://doi.org/10.1038/s12276-024-01328-6
CWAS-Plus: A new version of Category-Wide Association Study
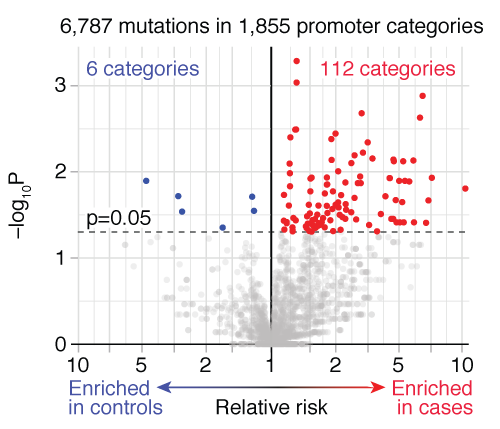
A new Python package runs category-wide association study with better user-interface. Now you can use this with pip install!
CWAS: Category-Wide Association Study
The Cython wrapper script runs category-wide association study presented in An et al. (2018) Science. This is an Amazon Machine Images (AMI) on Amazon Web Services (AWS) Cloud Computing Services.
De novo Risk Score and Annotation Clustering
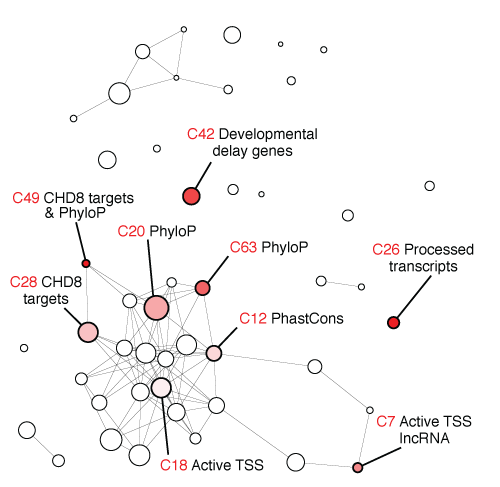
This repository contains R scripts for risk score analysis and annotation clustering in An et al. (2018) Science.
CatBurden: Category-based burden test
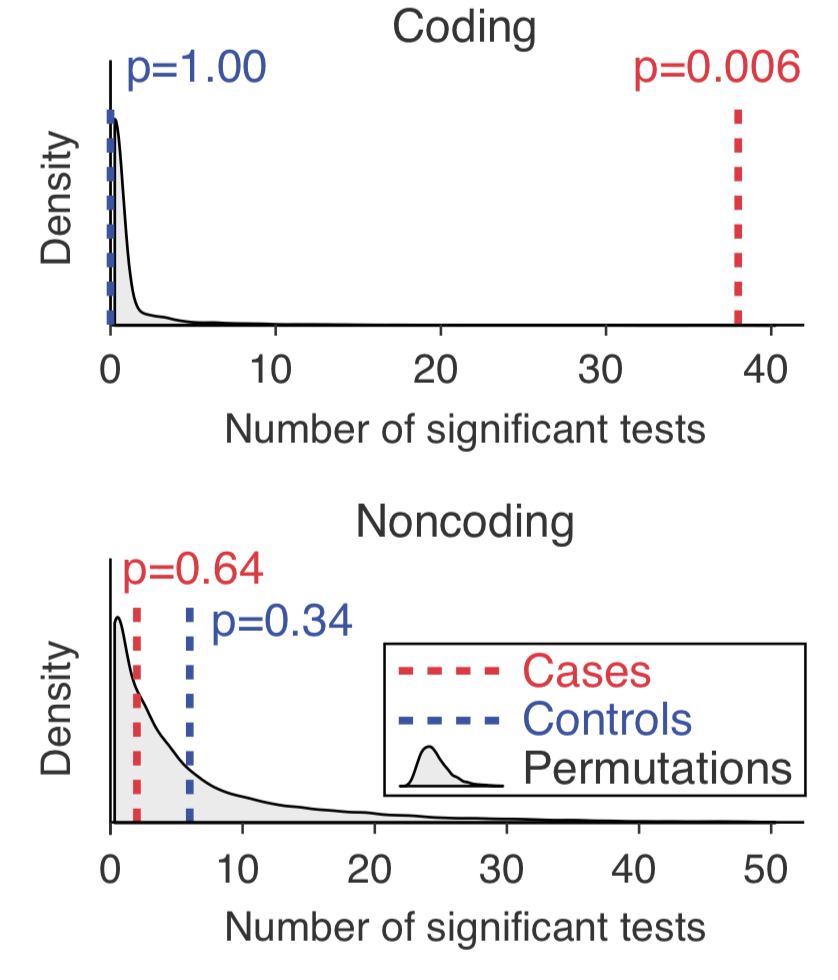
The R/Python wrapper script includes commands for the category-based burden test and the first edition of CWAS in the Werling et al. (2018) Nature Genetics. The scripts and materials are distributed in AMI (ID: ami-1eefb064) and you can deploy the workflow using AWS. We recommend using the 100 Gib size EBS and the EC2 instance type m4.large or m4.xlarge.
Hail tutorial (Korean)
우리말로 된 Hail 튜토리얼입니다.